Another publication in Nature describing the first de novo designed proteins with anti-cancer activity
Message boards : News : Another publication in Nature describing the first de novo designed proteins with anti-cancer activity
| Author | Message |
|---|---|
|
Admin Project administrator Send message Joined: 1 Jul 05 Posts: 5146 Credit: 0 RAC: 0 |
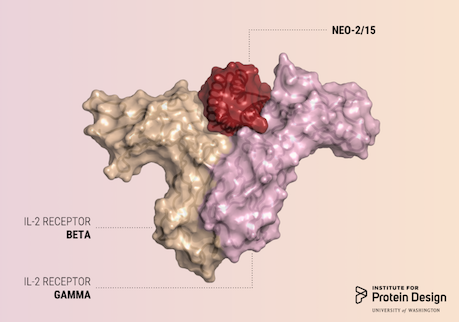 A report was published in Nature last week describing the first de novo designed proteins with anti-cancer activity. These compact molecules were designed to stimulate the same receptors as IL-2, a powerful immunotherapeutic drug, while avoiding unwanted off-target receptor interactions. We believe this is just the first of many computer-generated cancer drugs with enhanced specificity and potency. R@h participants provided computing for forward folding experiments used in this study which helped validate designs. We'd like to congratulate and thank all R@h volunteers who contributed to this work! Thank you! Read the full article here: https://www.nature.com/articles/s41586-018-0830-7 (PDF) |
|
biodoc Send message Joined: 19 Feb 06 Posts: 14 Credit: 30,720,744 RAC: 0 |
Congratulations! Unfortunately I'd have to pay for the article to read it but based on the abstract, it sounds very cool. |
![View the profile of [VENETO] boboviz Profile](https://boinc.bakerlab.org/rosetta/img/head_20.png) [VENETO] boboviz [VENETO] bobovizSend message Joined: 1 Dec 05 Posts: 2155 Credit: 12,998,581 RAC: 8,218 |
Congratulations! Unfortunately I'd have to pay for the article to read it but based on the abstract, it sounds very cool. In the news there is the link to complete pdf... |
|
steffen_moeller Send message Joined: 31 Oct 05 Posts: 2 Credit: 18,932,513 RAC: 0 |
I read through the acknowledgement section in the paper and searched for "home" in the article but failed to find any pointers to R@H. Did I not look carefully enough? I think this would be important for the project and for BOINC at large in many ways. |
|
Dad's computer Send message Joined: 22 Aug 19 Posts: 2 Credit: 935,495 RAC: 0 |
This is fantastic research. I have read many articles about promising drug discoveries. It seems that many wonderful discoveries are published in esteemed journals all over the world but then and nothing seems to happen. I have lost family and friends who have been treated with drugs found 50 years ago, drugs which I gather would not now pass drug safety testing but is all we have. I have recently seen a Ted lecture by an oncologist who says that despite the many billions of dollars spent world-wide on cancer research there has been no real progress in that 50 years. As is shown by the passion of the R@h community to help, we want to support you to make a fantastic paradigm shift from stone-age medicine, but how how quickly will this fantastic work be translated into therapies at the bedside? For those poor souls at the end of their treatment line there is no time to wait! |
![View the profile of [VENETO] boboviz Profile](https://boinc.bakerlab.org/rosetta/img/head_20.png) [VENETO] boboviz [VENETO] bobovizSend message Joined: 1 Dec 05 Posts: 2155 Credit: 12,998,581 RAC: 8,218 |
I have recently seen a Ted lecture by an oncologist who says that despite the many billions of dollars spent world-wide on cancer research there has been no real progress in that 50 years. I don't know who is this oncologist, but he said a lie. Childhood tumors, for example, now have 5ys survival rate over 80%, when 30 years ago was less than 30% |
 kaancanbaz kaancanbazSend message Joined: 28 Apr 06 Posts: 23 Credit: 3,045,052 RAC: 0 |
It’s good to see folding really makes difference. Hope to see more anti cancer discoveries from rah. |
|
YAG Send message Joined: 13 Oct 19 Posts: 7 Credit: 13,023,731 RAC: 10 |
It would be nice to announce this promising new in the new widget of the Rosseta@Home landing page, https://boinc.bakerlab.org/rosetta. The last published new is «The Audacious Project», 16 Jul 2019, 22:18:18 UTC. Have a nice day, and happy crunching! |
|
dadriano Volunteer moderator Project developer Project scientist Send message Joined: 22 Jan 15 Posts: 3 Credit: 347,742 RAC: 0 |
Dear steffen_moller: What a massive oversight from me! This is Daniel-Adriano Silva, author in the paper. You are entirely right. I spend a long time in disbelief looking for the Rosetta@Home acknowledgment in the article (it should have been there), and it is missing. I am sorry, please accept my apology and let me elaborate: In figure #1 panel "e", the bottom panel shows the use of "Forward Folding" and ("Fast Forward Folding", also known as biased Forward Folding, see Marcos et. al., https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6219906/) to filter/evaluate the computational designs. We executed many of these simulations for thousands of computational designs, and many of these run on BIONC's Rosetta@Home. The legend of Fig.1 should (instead) read as: ... e) Top: Rosetta flexible backbone sequence design; Bottom: Forward Folding simulations for filtering computational-models with smooth folding funnels (see Methods)... The methods and/or acknowledgments do mention the use of multiple computational resources. However, they should have also included R@H, but it seems that we/I accidentally miss it. I apologize. I hope this clarifies that R@H did contribute to this development. Please receive my acknowledgments, and know that we value your important contribution that helps us bring de novo protein to solve real-world problems. Best, Daniel |
 yoerik yoerikSend message Joined: 24 Mar 20 Posts: 128 Credit: 169,525 RAC: 0 |
Dear steffen_moller: What a massive oversight from me! This is Daniel-Adriano Silva, author in the paper. Just have to say, Daniel - wow. It takes a large amount of guts and morals to admit a wrong - especially when you could've easily ignored this and let it slide. Thank you for your hard work, and thank you for taking the time to share this with us. You're not just working for a university or this project - you're working for humanity - and I just need to say that I genuinely appreciate it. Keep up the good work. |
|
Falconet Send message Joined: 9 Mar 09 Posts: 355 Credit: 1,669,337 RAC: 14 |
Read this thread, especially the links: https://boinc.bakerlab.org/rosetta/forum_thread.php?id=14222 Neoleukin-2/15 will be entering human clinical trials either in late 2020 or mid-2021. Hopefully, Rosetta@home will publish an update on this. |
|
Falconet Send message Joined: 9 Mar 09 Posts: 355 Credit: 1,669,337 RAC: 14 |
Neoleukin Therapeutics Announces Submission of Investigational New Drug Application for NL-201 De Novo Protein Immunotherapy Candidate for Cancer "SEATTLE, Dec. 10, 2020 (GLOBE NEWSWIRE) -- Neoleukin Therapeutics, Inc., “Neoleukin” (NASDAQ:NLTX), a biopharmaceutical company utilizing sophisticated computational methods to design de novo protein therapeutics, today announced the submission of an Investigational New Drug (IND) Application with the U.S. Food and Drug Administration to begin a Phase 1 clinical program of its lead immunotherapeutic candidate, NL-201. NL-201 is a computationally designed de novo protein that is a mimetic of natural cytokines IL-2 and IL-15." "The Phase 1 study is expected to enroll up to 120 patients with relapsed or refractory solid tumors. Patients will receive intravenous monotherapy with NL-201 in order to assess safety, pharmacokinetics, pharmacodynamics, and antitumor activity. When the recommended dose and schedule are determined, indication-specific expansion cohorts will be enrolled to estimate safety and antitumor activity. In addition to the IND application, Neoleukin has submitted a Clinical Trial Notification (CTN) application for NL-201 in Australia." |
 rochester new york rochester new yorkSend message Joined: 2 Jul 06 Posts: 2842 Credit: 2,020,043 RAC: 0 |
|
Message boards :
News :
Another publication in Nature describing the first de novo designed proteins with anti-cancer activity

©2026 University of Washington
https://www.bakerlab.org